.jpg)
MOHAMMAD IMRAN HOSSAIN
Email: hossainimran.maia@gmail.com
Address: 1 Place Montespan, 91380 Chilly-Mazarin, France.
Please Click to [Download CV]
Copyright © Mohammad Imran Hossain Official Webpage 2024.
About Me
I am a graduate student of the Master of Computer Science (Bioinformatics and Modeling specialization) at the Sorbonne University in France.
I priviously completed prestigious Erasmus Mundus Joint Master Degree in Medical Imaging and Applications,
fully funded by the European Union and jointly coordinated by the
University of Girona (Spain),
the University of Cassino and Southern Lazio (Italy), and
the University of Burgundy (France).
Additionally, I was also a recipient of the
Stipendium Hungaricum Scholarship for
the Master of Science in Electrical Engineering at the
Budapest University of Technology and Economics (Hungary).
I received the Bachelor of Science in Electrical and Electronic Engineering with the highest academic distinction 'Summa Cum Laude' from the United International University (Bangladesh) in 2022. My undergraduate research project "Electronic Toll Collection System Using Optical Wireless Communication"
was supervised by Professor Dr. Raqibul Mostafa.
Specializing in AI-driven Medical Image Analysis and Computer-Aided Diagnosis (CAD), I am committed to advancing healthcare.
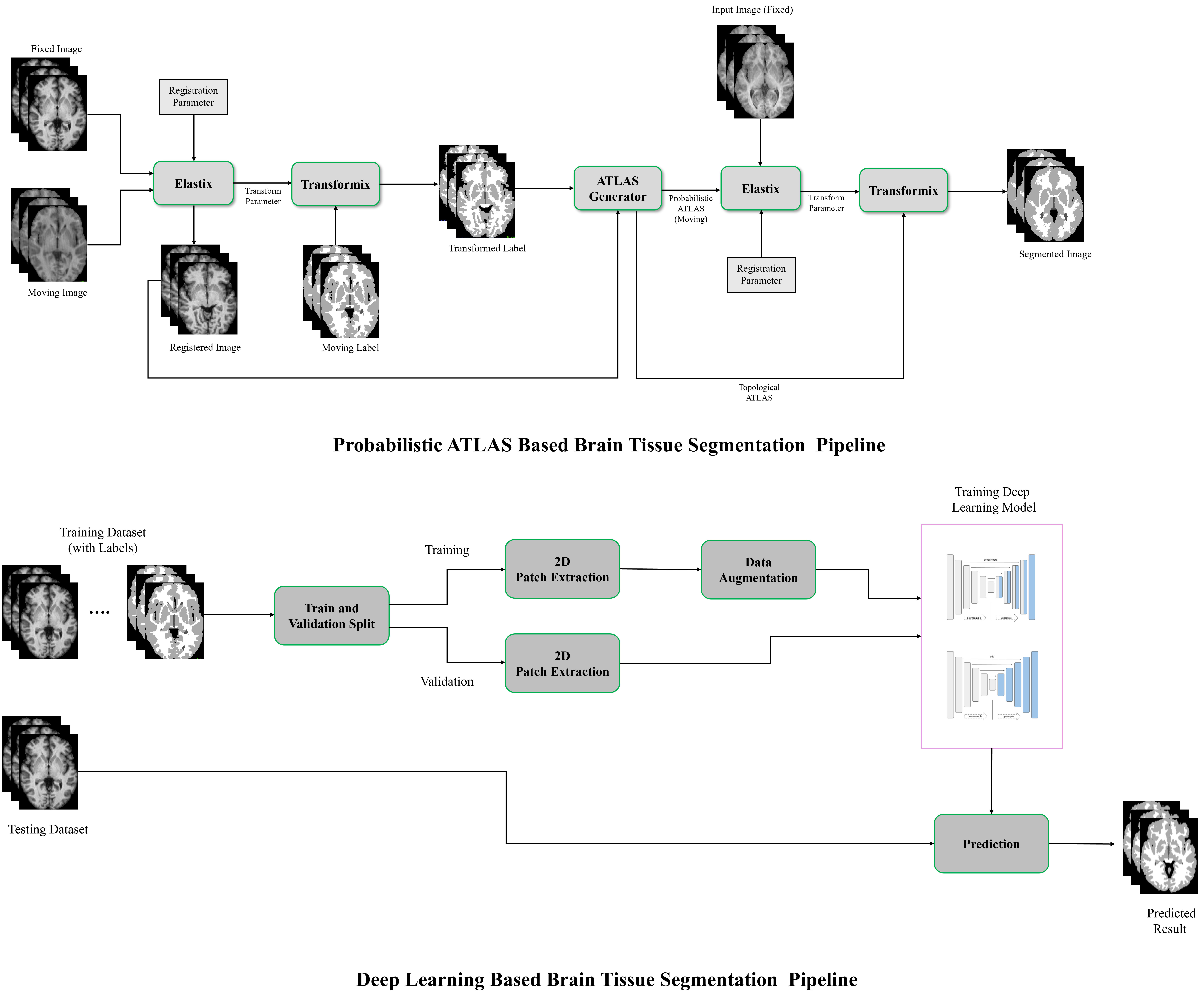
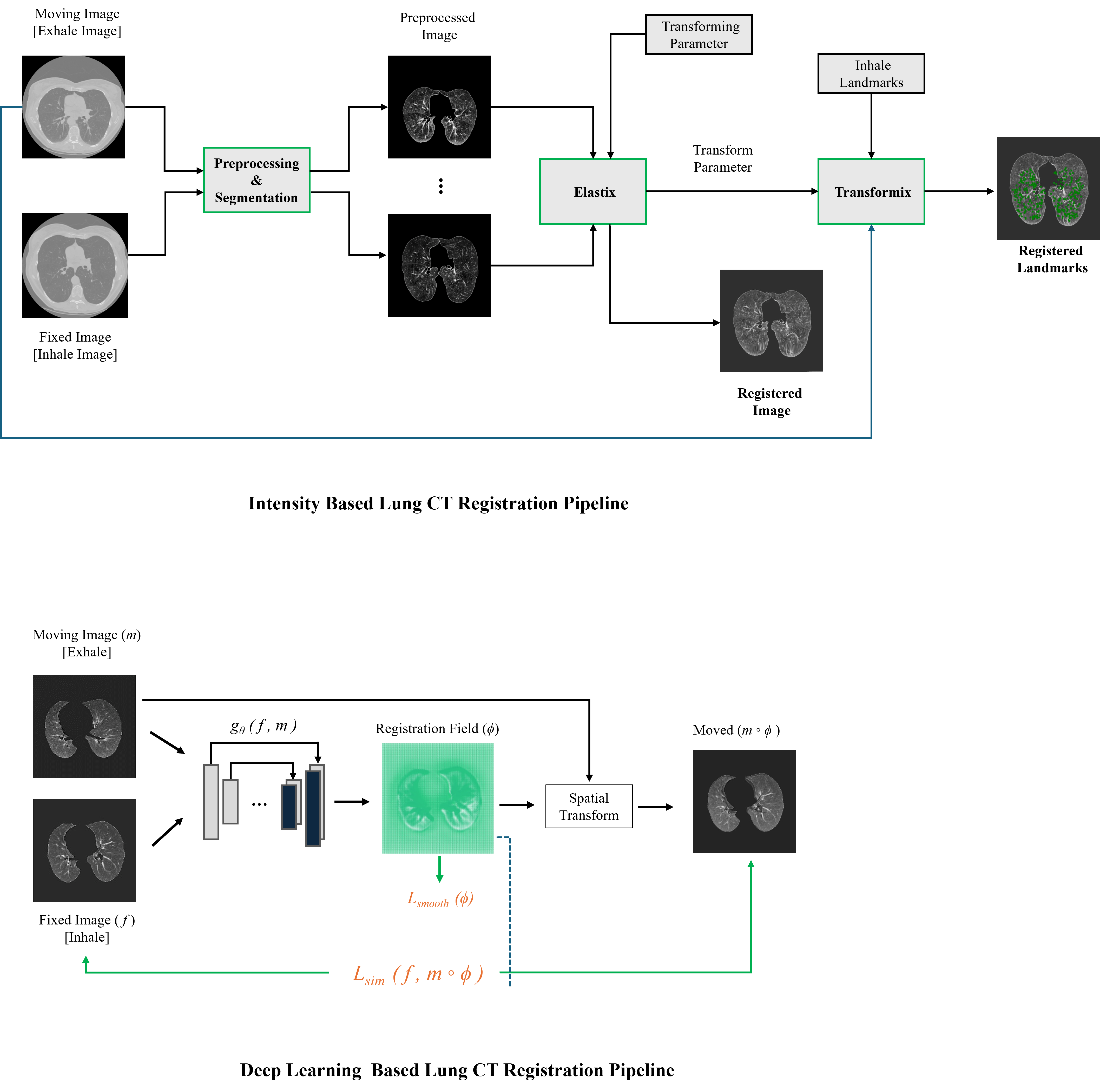
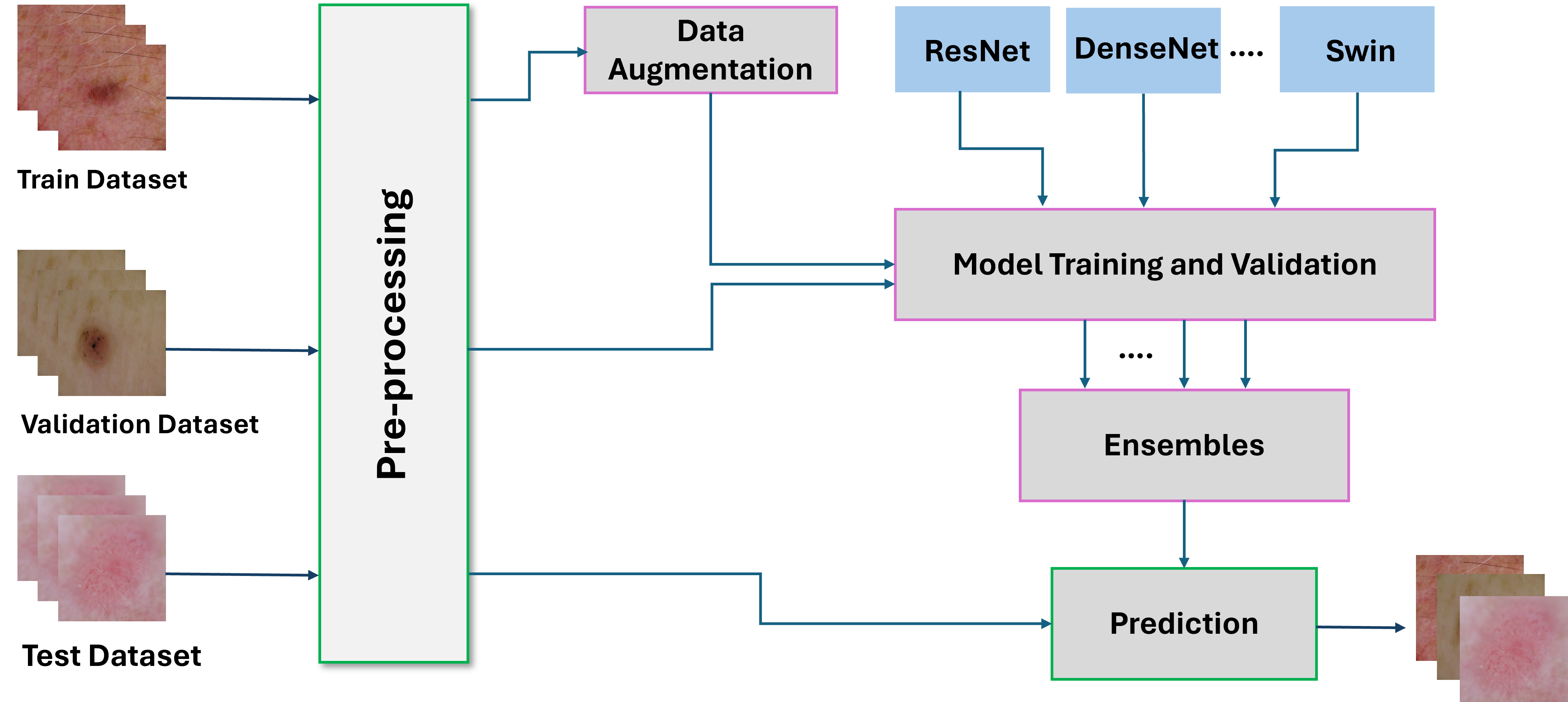
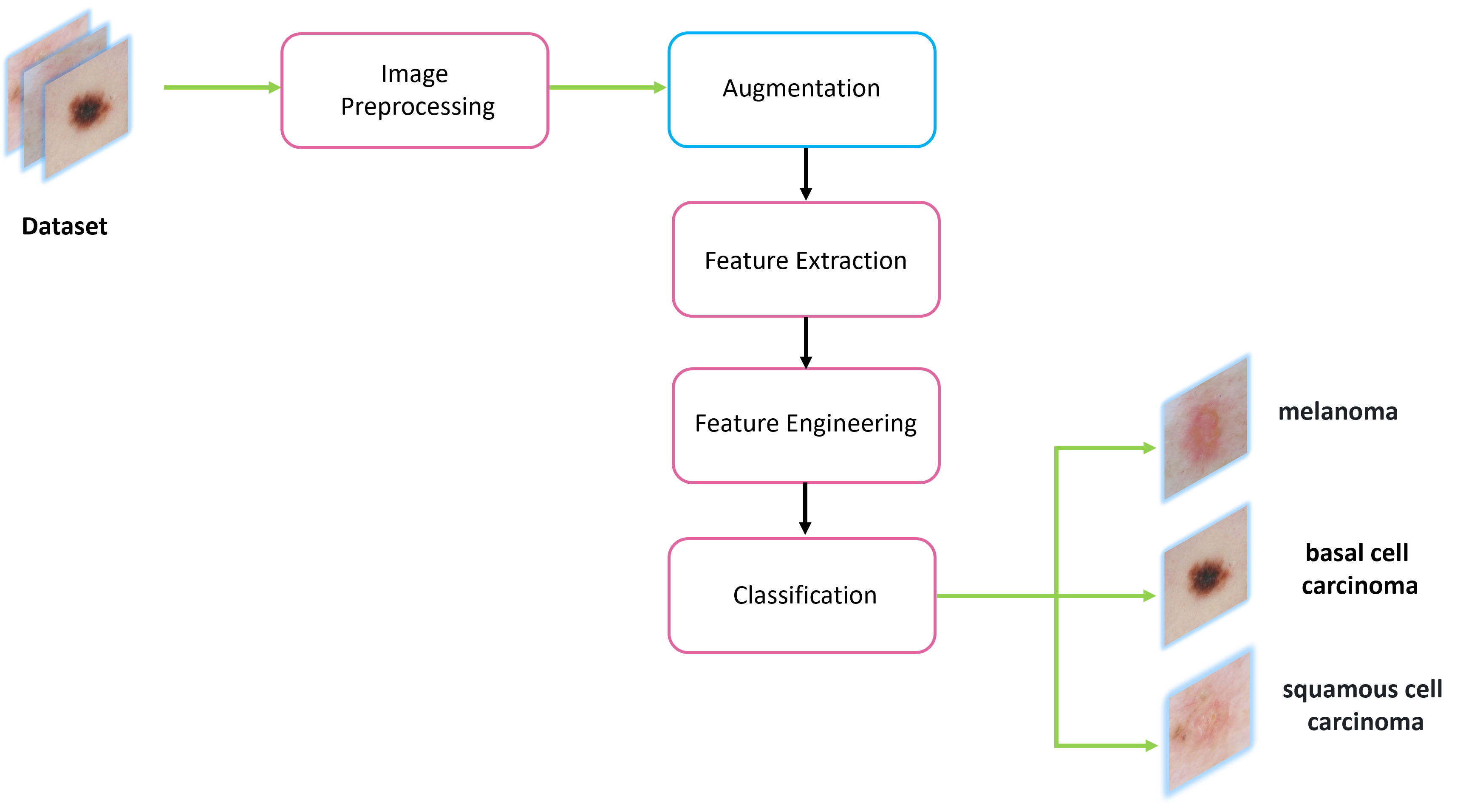
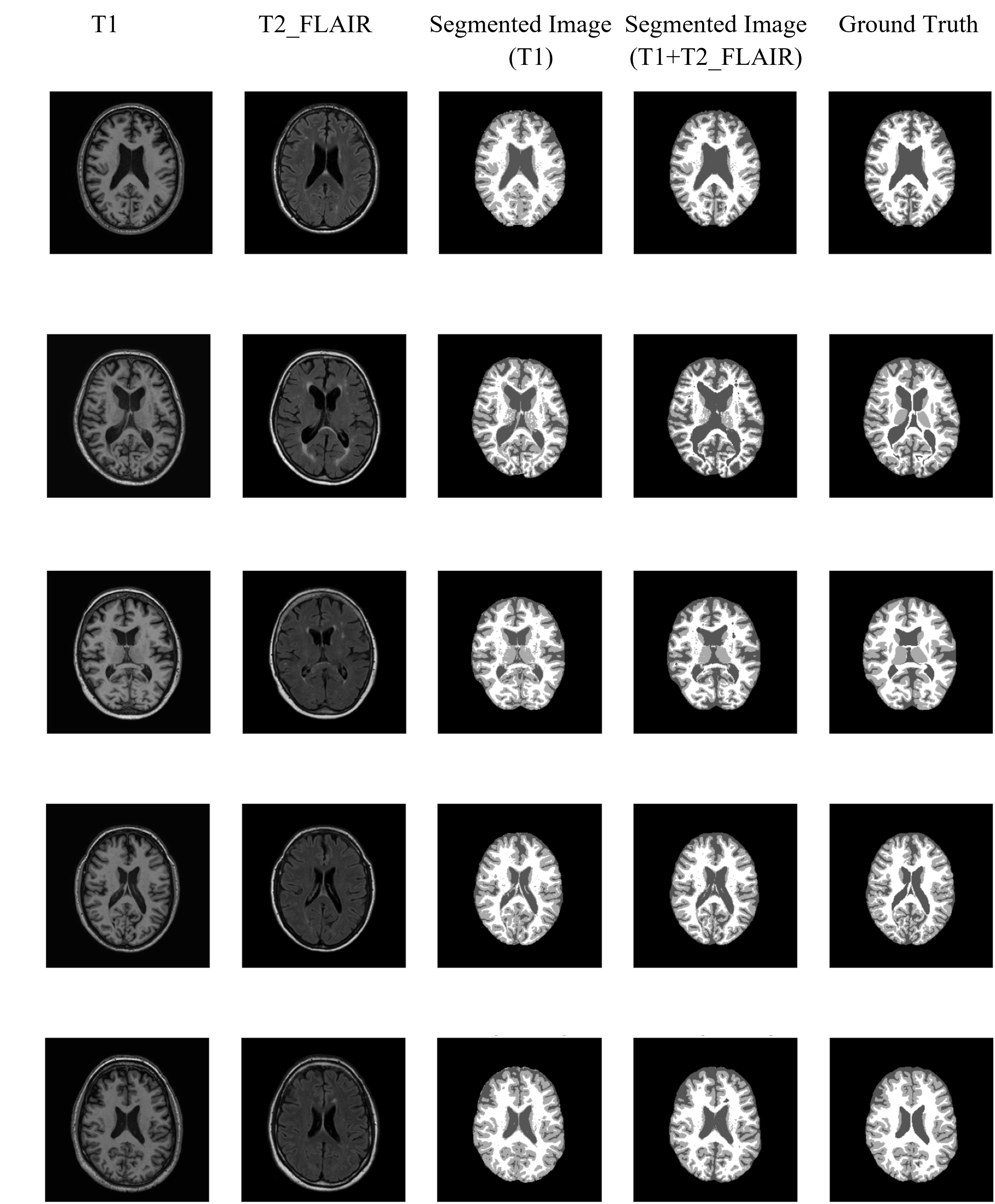
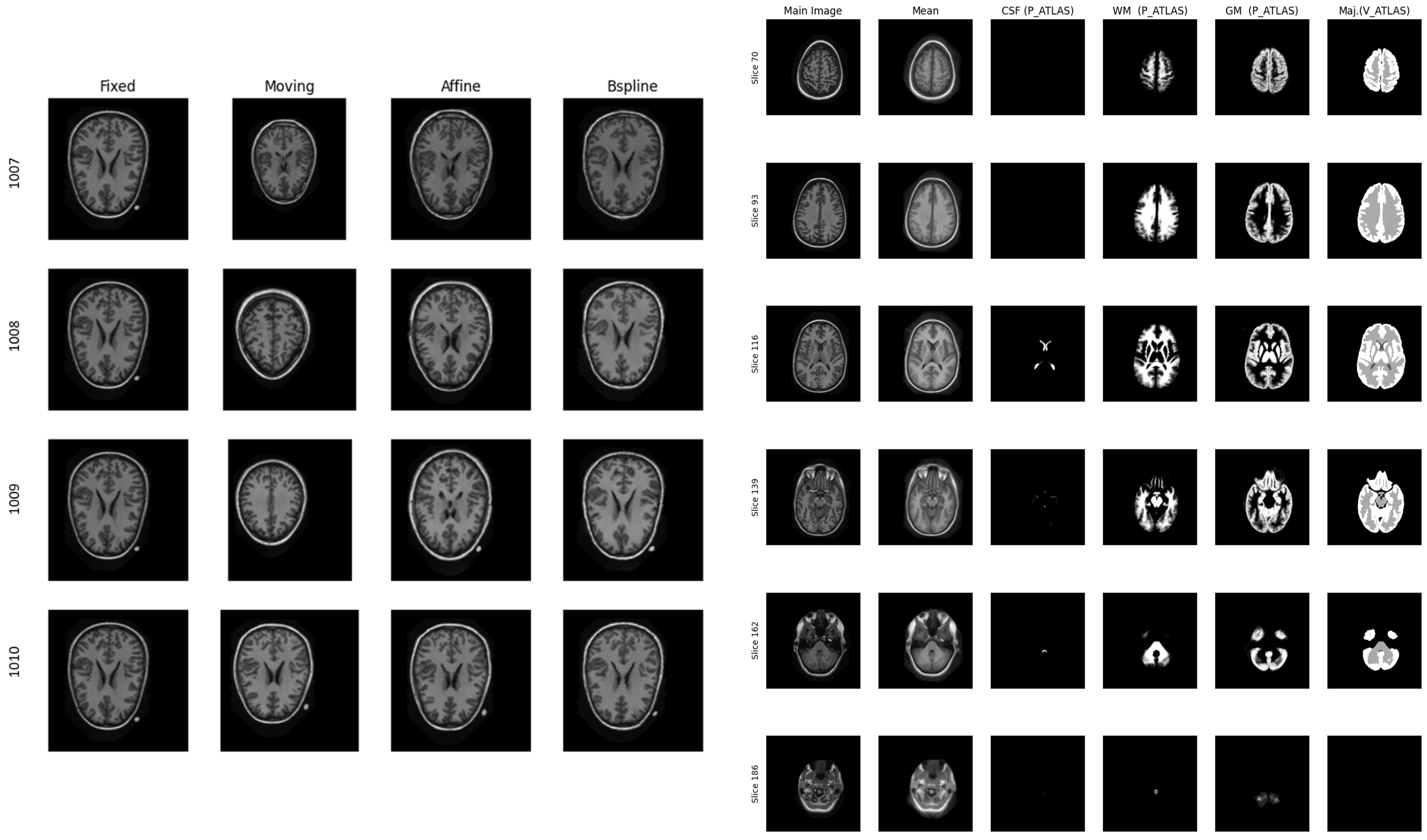
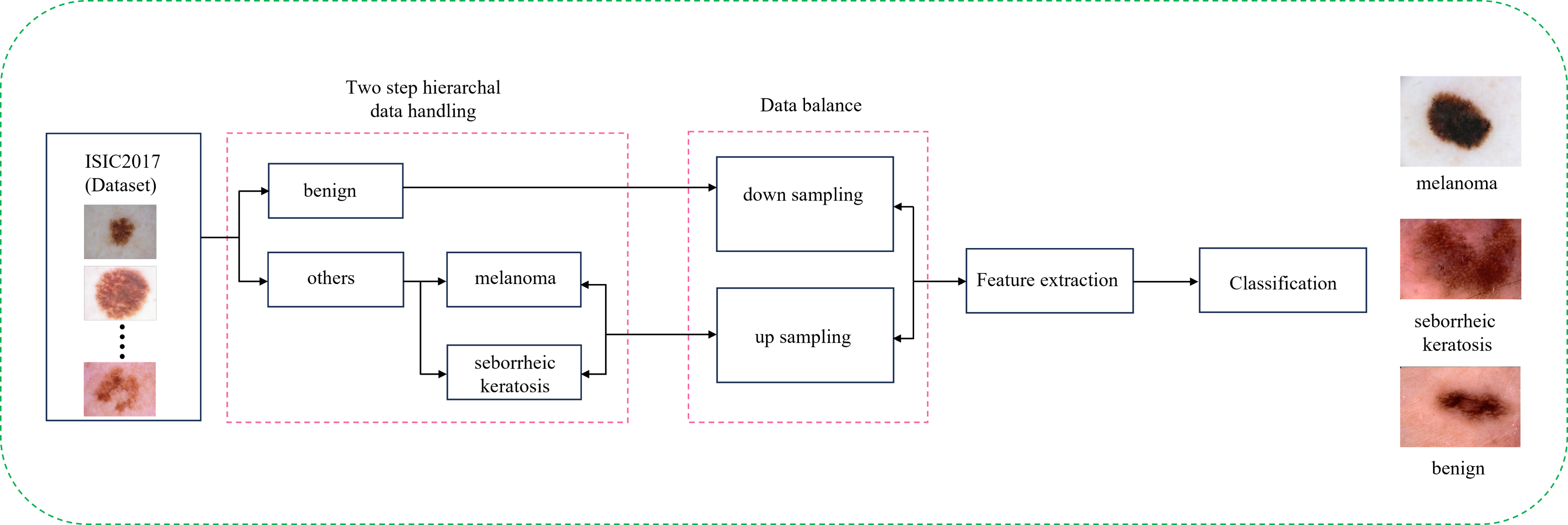
My research projects include Skin Lesion Detection, Functional Neuroimaging, Brain Tissue Registration and Segmentation,
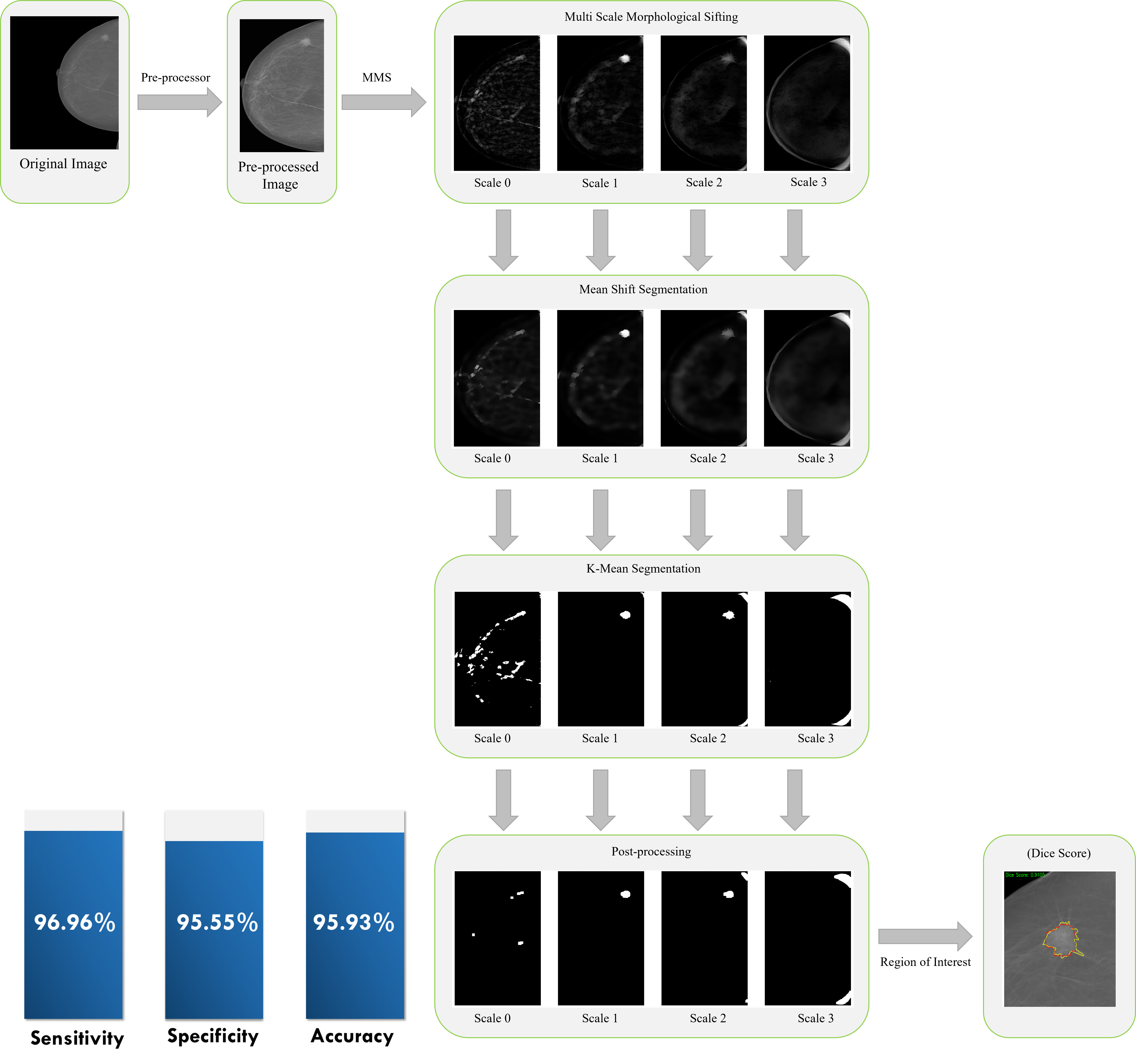
Alzheimer's Disease Classification, Breast Mass Segmentation and Detection, and Computational Pathology.
I aim to contribute innovative solutions for accurate diagnosis and improved patient outcomes.
I completed my master's internship at the National Center for Scientific Research (CNRS)
in France, working on the project titled "Deep Learning-Based Detection of Homologous Recombination Deficiency (HRD) in Breast and Ovarian Cancer Whole Slide Histopathology Images" with
Professor Dr. Manon Ansart.
Before that, I worked as a visiting researcher at the Diagnostic Image Analysis Group (DIAG) at Radboud University Medical Center in
The Netherlands on the project "Real-time MR Image Reconstruction Using Deep Learning" under the supervision of Stan Noordman.
I am driven by a commitment to advancing healthcare through innovative solutions and look forward to contributing significantly to
the intersection of engineering and medicine in my future endeavours.
Research Interests: Medical Image Analysis & Computation, Computational Pathology, Computer-Aided Diagnosis, Machine & Deep Learning, AI in HealthCare.
Important Updates:
- [Feb 2024] Started Master's Thesis at the National Center for Scientific Research [CNRS] (France).
- [Jan 2024] Ranked 2nd in the MAIA Brain Tissue Segmentation Challenge at UdG (Spain).
- [Nov 2023] Ranked 2nd in the MAIA CADx Skin Lesion Classification Challenge at UdG (Spain).
- [Sep 2023] Started the third semester of the Joint Master's at the Univeristy of Girona (Spain).
- [Sep 2023] Participated in the 17th EXCITE Summer School on Biomedical Imaging at UZH and ETH Zurich (Switzerland).
- [Jul 2023] Started summer internship at DIAG, RadboudUMC (The Netherlands).
- [Jun 2023] Breast mass detection research project is presented at the Advance Image Analysis project defence at UNICAS (Italy).
- [Jun 2023] Skin lesion detection research project is presented at the Machine & Deep Learning project defence at UNICAS (Italy).
- [Jun 2023] Ranked 2nd in the regression challenge at the Italian National Research Council (Italy).
- [Feb 2023] Started the second semester of the Joint Master's at the Univeristy of Cassino (Italy).
- [Jan 2023] Presented research poster on Parkinson's disease and EEG signals at uB (France).
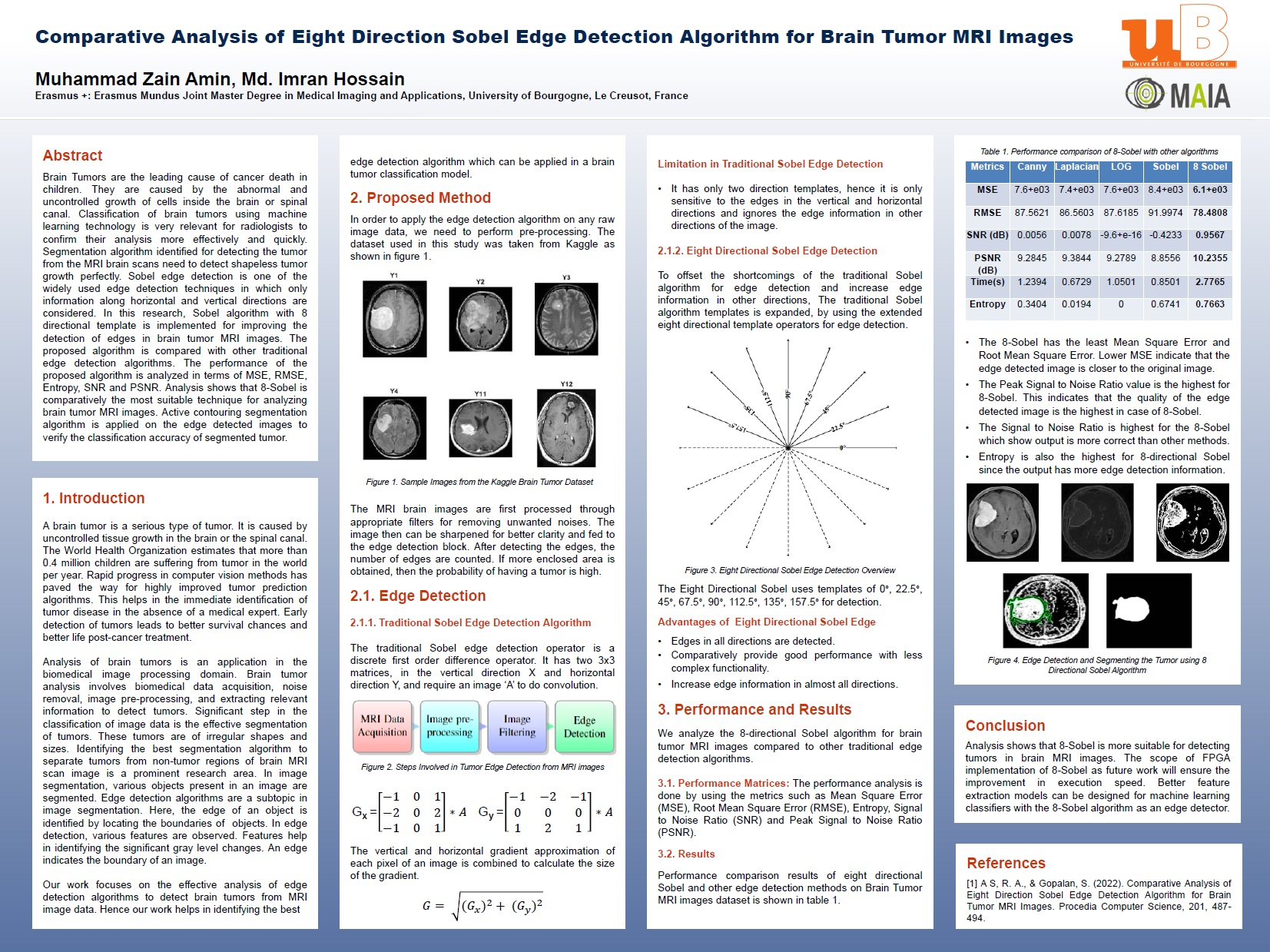
- [Jan 2023] Presented research poster on Brain Tumor detection from MR Images at uB (France).
- [Dec 2022] Bibliography review project on Brain Tumor Detection is presented at the Medical Sensor project defence at uB (France).
- [Sep 2022] Started the first semester of the Joint Master's at the Univeristy of Burgundy (France).
Ongoing Research:
- Deep Learning-Based Detection of Homologous Recombination Deficiency (HRD) in Breast and Ovarian Cancer Whole Slide Histopathology Images.
- Identification of Osteonecrosis using Artificial Intelligence and Machine Learning.